RandomizedSearchCV#
A diferencia de GridSearchCV, RandomizedSearchCV no prueba con
todos los valores de los hiperparámetros, sino que se muestrea un número
fijo de configuraciones de hiperparámetros.
El número de configuraciones de hiperparámetros que se prueban se
especifica con el argumento n_iter=.
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
from sklearn.model_selection import train_test_split
from sklearn.preprocessing import StandardScaler
from keras.models import Sequential
from keras.layers import Dense
import keras
import warnings # Para ignorar mensajes de advertencia
warnings.filterwarnings("ignore")
df = pd.read_csv("spiral.csv", sep=",", decimal=".")
print(df.head())
X1 X2 y
0 13.31596 -5.47317 0.0
1 16.70656 1.26700 0.0
2 7.18348 7.29934 1.0
3 -7.94940 -14.77444 1.0
4 -5.96108 -18.68620 1.0
plt.scatter(df.iloc[:,0],df.iloc[:,1], c = df.iloc[:,2])
plt.show()

Conjunto de train y test:#
X = pd.concat([df[["X1", "X2"]]], axis=1)
print(X)
X1 X2
0 13.31596 -5.47317
1 16.70656 1.26700
2 7.18348 7.29934
3 -7.94940 -14.77444
4 -5.96108 -18.68620
... ... ...
3995 4.67165 11.72679
3996 -9.34366 0.81879
3997 -13.08116 -14.54117
3998 10.66048 -17.28003
3999 12.35733 -8.63444
[4000 rows x 2 columns]
y = df["y"]
print(y)
0 0.0
1 0.0
2 1.0
3 1.0
4 1.0
...
3995 1.0
3996 0.0
3997 1.0
3998 1.0
3999 0.0
Name: y, Length: 4000, dtype: float64
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.2, random_state=0)
Escalado de variables:#
sc = StandardScaler()
sc.fit(X_train)
X_train = sc.transform(X_train)
X_test = sc.transform(X_test)
Optimización de neuronas y epochs:#
Construcción de la red neuronal:
def create_model(units = 1):
model = Sequential()
model.add(Dense(units, activation = "relu", input_shape=(X.shape[1],)))
model.add(Dense(units, activation = "relu"))
model.add(Dense(1, activation = "sigmoid"))
model.compile(loss = "binary_crossentropy", optimizer = "adam", metrics = ["accuracy"])
return model
Hiperparámetros a ajustar:
Creamos una lista de hiperparámetros y de los valores que cada uno tendrá.
param_grid = dict(units = [2, 4, 6, 8], epochs = [10, 20, 30, 50, 80])
param_grid
{'units': [2, 4, 6, 8], 'epochs': [10, 20, 30, 50, 80]}
Uso de la API de Scikit-Learn sobre Keras:
from keras.wrappers.scikit_learn import KerasClassifier
keras_reg = KerasClassifier(create_model, verbose = 1)
RandomizedSearchCV de Scikit-Learn para optimizar hiperparámetros:
from sklearn.model_selection import RandomizedSearchCV
En RandomizedSearchCV el argumento para indicar las posibles
combinaciones de hiperparámetros se pasa con param_distributions=.
El número de configuraciones de hiperparámetros que se probarán se
especifica con el argumento n_iter=.
Tenemos en param_grid cuatro configuraciones para units y cinco
configuraciones para epochs.
Si en RandomizedSearchCV indicamos n_iter=3, de cada
hiperparámetro seleccionará tres configuraciones y con cv=2
realizará el ajuste dos veces con subconjuntos de datos aleatorios
(cross-validation), por tanto, entrena la red neuronal seis veces.
Por defecto n_iter=10 y cv=5.
random_search = RandomizedSearchCV(estimator=keras_reg, param_distributions=param_grid, n_iter=3, cv=2)
random_search.fit(X_train, y_train,
validation_data = (X_test, y_test),
verbose = 0)
50/50 [==============================] - 0s 956us/step - loss: 0.5424 - accuracy: 0.6625
50/50 [==============================] - 0s 956us/step - loss: 0.5451 - accuracy: 0.6950
50/50 [==============================] - 0s 637us/step - loss: 0.6171 - accuracy: 0.6438
50/50 [==============================] - 0s 1ms/step - loss: 0.6098 - accuracy: 0.6650
50/50 [==============================] - 0s 638us/step - loss: 0.3781 - accuracy: 0.8250
50/50 [==============================] - 0s 743us/step - loss: 0.3598 - accuracy: 0.8487
RandomizedSearchCV(cv=2,
estimator=<keras.wrappers.scikit_learn.KerasClassifier object at 0x000001F978F035B0>,
n_iter=3,
param_distributions={'epochs': [10, 20, 30, 50, 80],
'units': [2, 4, 6, 8]})
Configuraciones de hiperparámetros seleccionadas al azar:
Los hiperparámetros seleccionados al azar de acuerdo con lo pasado en el
argumento n_iter se muestran a continuación.
random_search.cv_results_["params"]
[{'units': 4, 'epochs': 50},
{'units': 8, 'epochs': 10},
{'units': 6, 'epochs': 80}]
Mejor modelo:
random_search.best_params_
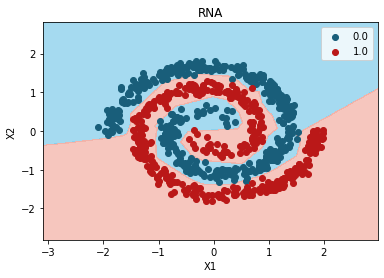
{'units': 6, 'epochs': 80}
Predicción con el mejor modelo:
y_pred = random_search.best_estimator_.predict(X_test, verbose = 0)
y_pred[0:5]
array([[0.],
[1.],
[0.],
[1.],
[0.]])
from matplotlib.colors import ListedColormap
X_Set, y_Set = X_test, y_test
X1, X2 = np.meshgrid(
np.arange(start=X_Set[:, 0].min() - 1, stop=X_Set[:, 0].max() + 1, step=0.01),
np.arange(start=X_Set[:, 1].min() - 1, stop=X_Set[:, 1].max() + 1, step=0.01),
)
plt.contourf(
X1,
X2,
random_search.best_estimator_.predict(np.array([X1.ravel(), X2.ravel()]).T).reshape(X1.shape),
alpha=0.75,
cmap=ListedColormap(("skyblue", "#F3B3A9"))
)
plt.xlim(X1.min(), X1.max())
plt.ylim(X2.min(), X2.max())
for i, j in enumerate(np.unique(y_Set)):
plt.scatter(
X_Set[y_Set == j, 0],
X_Set[y_Set == j, 1],
c=ListedColormap(("#195E7A", "#BA1818"))(i),
label=j,
)
plt.title("RNA")
plt.xlabel("X1")
plt.ylabel("X2")
plt.legend()
plt.show()
10715/10715 [==============================] - 6s 582us/step
c argument looks like a single numeric RGB or RGBA sequence, which should be avoided as value-mapping will have precedence in case its length matches with x & y. Please use the color keyword-argument or provide a 2D array with a single row if you intend to specify the same RGB or RGBA value for all points. c argument looks like a single numeric RGB or RGBA sequence, which should be avoided as value-mapping will have precedence in case its length matches with x & y. Please use the color keyword-argument or provide a 2D array with a single row if you intend to specify the same RGB or RGBA value for all points.
Optimización de neuronas, capas ocultas, función de activación, optimizadores, batch y epochs:#
Construcción de la red neuronal:
def create_model(units = 1, n_hidden = 2, activation = "relu", optimizer = "adam"):
model = Sequential()
# Dimensión de las entradas para la primera capa:
model.add(keras.layers.InputLayer(input_shape=(X.shape[1],)))
# Loop para las capas ocultas:
for layer in range(n_hidden):
model.add(Dense(units, activation=activation))
model.add(Dense(1, activation = "sigmoid"))
model.compile(loss = "binary_crossentropy", optimizer = optimizer, metrics = ["accuracy"])
return model
Hiperparámetros a ajustar:
param_grid = dict(units = [2, 4, 6, 8],
n_hidden = [2, 3],
activation = ["relu", "tanh", "sigmoid"],
optimizer = ["adam", "rmsprop"],
batch_size = [10, 100],
epochs = [100, 200, 300])
param_grid
{'units': [2, 4, 6, 8],
'n_hidden': [2, 3],
'activation': ['relu', 'tanh', 'sigmoid'],
'optimizer': ['adam', 'rmsprop'],
'batch_size': [10, 100],
'epochs': [100, 200, 300]}
Uso de la API de Scikit-Learn sobre Keras:
keras_reg = KerasClassifier(create_model, verbose = 0)
GridSearchCV de Scikit-Learn para optimizar hiperparámetros:
random_search = RandomizedSearchCV(estimator=keras_reg, param_distributions=param_grid, n_jobs=-1, n_iter=10)
import time
StartTime = time.time()
random_search.fit(X_train, y_train,
validation_data = (X_test, y_test),
verbose = 0)
EndTime = time.time()
print("---------> Tiempo en procesar: ", round((EndTime - StartTime) / 60), "Minutos")
---------> Tiempo en procesar: 12 Minutos
Configuraciones de hiperparámetros seleccionadas al azar:
random_search.cv_results_["params"]
[{'units': 2,
'optimizer': 'adam',
'n_hidden': 2,
'epochs': 100,
'batch_size': 10,
'activation': 'sigmoid'},
{'units': 6,
'optimizer': 'adam',
'n_hidden': 3,
'epochs': 200,
'batch_size': 100,
'activation': 'sigmoid'},
{'units': 8,
'optimizer': 'rmsprop',
'n_hidden': 2,
'epochs': 200,
'batch_size': 100,
'activation': 'sigmoid'},
{'units': 6,
'optimizer': 'rmsprop',
'n_hidden': 2,
'epochs': 300,
'batch_size': 10,
'activation': 'tanh'},
{'units': 8,
'optimizer': 'adam',
'n_hidden': 2,
'epochs': 200,
'batch_size': 10,
'activation': 'sigmoid'},
{'units': 8,
'optimizer': 'adam',
'n_hidden': 2,
'epochs': 300,
'batch_size': 10,
'activation': 'relu'},
{'units': 6,
'optimizer': 'rmsprop',
'n_hidden': 2,
'epochs': 200,
'batch_size': 100,
'activation': 'sigmoid'},
{'units': 6,
'optimizer': 'adam',
'n_hidden': 3,
'epochs': 100,
'batch_size': 100,
'activation': 'relu'},
{'units': 6,
'optimizer': 'rmsprop',
'n_hidden': 3,
'epochs': 100,
'batch_size': 100,
'activation': 'relu'},
{'units': 2,
'optimizer': 'adam',
'n_hidden': 2,
'epochs': 300,
'batch_size': 10,
'activation': 'relu'}]
Mejor modelo:
random_search.best_params_
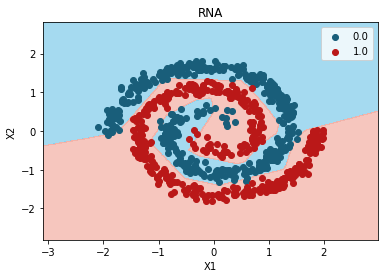
{'units': 8,
'optimizer': 'adam',
'n_hidden': 2,
'epochs': 300,
'batch_size': 10,
'activation': 'relu'}
Predicción con el mejor modelo:
y_pred = random_search.best_estimator_.predict(X_test, verbose = 0)
y_pred[0:5]
array([[0.],
[1.],
[0.],
[1.],
[0.]])
from matplotlib.colors import ListedColormap
X_Set, y_Set = X_test, y_test
X1, X2 = np.meshgrid(
np.arange(start=X_Set[:, 0].min() - 1, stop=X_Set[:, 0].max() + 1, step=0.01),
np.arange(start=X_Set[:, 1].min() - 1, stop=X_Set[:, 1].max() + 1, step=0.01),
)
plt.contourf(
X1,
X2,
random_search.best_estimator_.predict(np.array([X1.ravel(), X2.ravel()]).T).reshape(X1.shape),
alpha=0.75,
cmap=ListedColormap(("skyblue", "#F3B3A9")), verbose = 0
)
plt.xlim(X1.min(), X1.max())
plt.ylim(X2.min(), X2.max())
for i, j in enumerate(np.unique(y_Set)):
plt.scatter(
X_Set[y_Set == j, 0],
X_Set[y_Set == j, 1],
c=ListedColormap(("#195E7A", "#BA1818"))(i),
label=j,
)
plt.title("RNA")
plt.xlabel("X1")
plt.ylabel("X2")
plt.legend()
plt.show()
10715/10715 [==============================] - 7s 620us/step
c argument looks like a single numeric RGB or RGBA sequence, which should be avoided as value-mapping will have precedence in case its length matches with x & y. Please use the color keyword-argument or provide a 2D array with a single row if you intend to specify the same RGB or RGBA value for all points. c argument looks like a single numeric RGB or RGBA sequence, which should be avoided as value-mapping will have precedence in case its length matches with x & y. Please use the color keyword-argument or provide a 2D array with a single row if you intend to specify the same RGB or RGBA value for all points.